Imaging Biomarkers
We are developing a number of imaging biomarkers based on quantitative molecular imaging techniques to quantify spatio-temporal development of tumor heterogeneity and treatment resistance.
- Kinetic Modeling
- Radiomic Interrogation of Imaging Biomarkers
- Harmonization of PET Scanners
- Quantitative Total Bone Imaging
- Deep Learning
- Robust Optimization of Radiation Therapy
- Temporal evolution of radiotracer spatial distribution is more informative than the spatial distribution at single timepoint
- Dynamic acquisition and kinetic modeling enable extraction of the information offered by particular radiotracer and allows for better image quantification
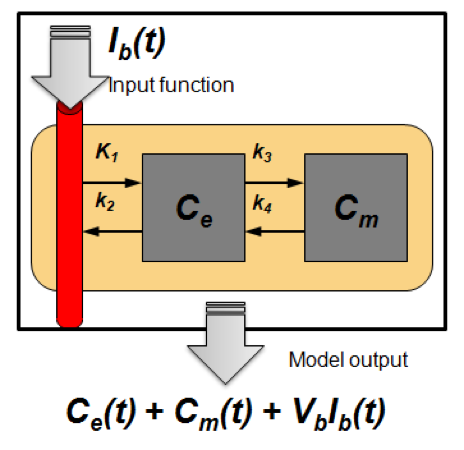
-
- Estimated model parameters are used for further analysis
- Vascular and proliferative flare show complimentary but distinct patterns
- Primarily used for FLT PET
- Methodology translated for DCE-CT
- Current research: regularization methods to make the parameter optimization more robust
-
- How do we identify imaging patterns correlated with clinical and biological properties?
- Convert imaging data to high dimensional mineable feature space (radiomics)
- We are exploring various radiomics-based initiatives to enhance treatment response assessment and link imaging with genetics
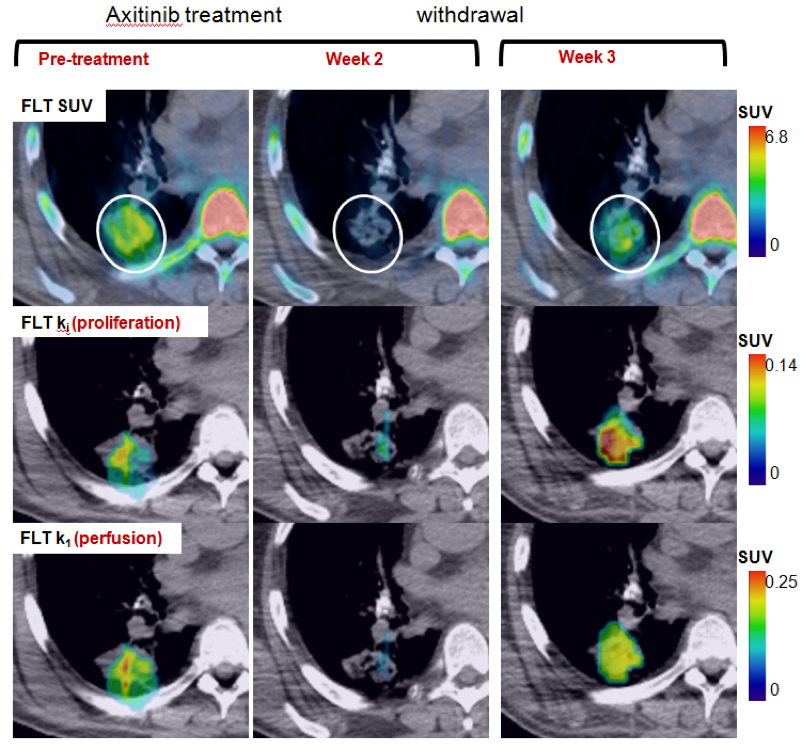
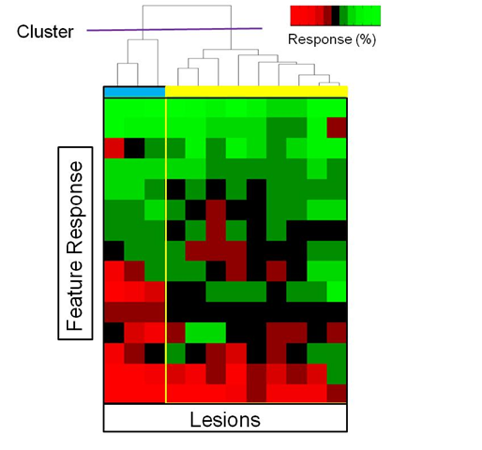 Subpopulations of Imaging Response:
Subpopulations of Imaging Response:
- Sub-grouping lesions to fully characterize responses to treatment
- Using MIB, different populations can be targeted for biopsy
- Research focus: quantitatively assessing imaging-genomic associations of drug resistance
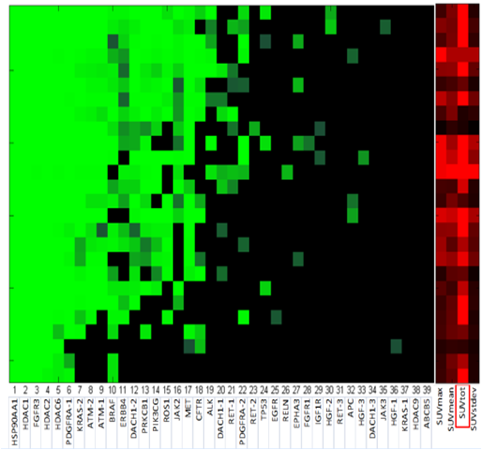 Cellular biomarkers of imaging resistance:
Cellular biomarkers of imaging resistance:
- Co-expression of genetic markers and imaging metrics for NSCLC.
- Radiogenomic models allow cellular data to be linked with the quantitative imaging
- Research focus: prognostic and predictive value of imaging features with disease type
-
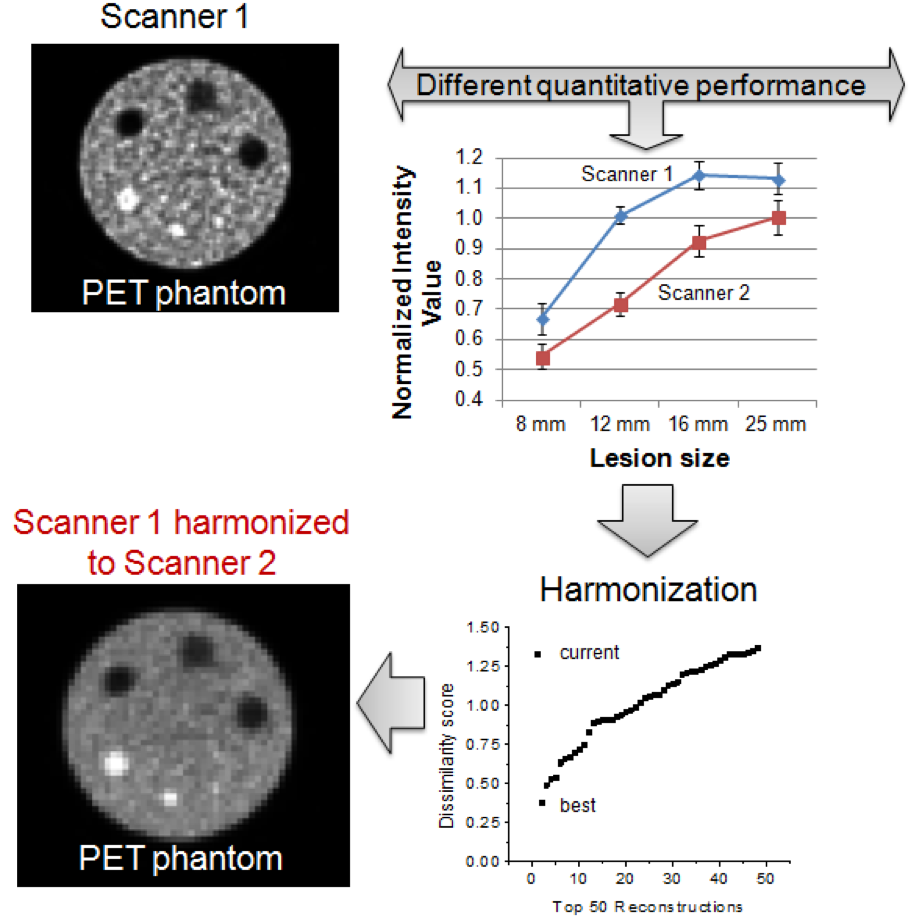

- Differences in scanner performance adds uncertainty to multi-center imaging clinical studies
- Scanners can be harmonized by finding reconstruction parameters that maximize similarities in images
- Harmonization of scanners increases the likelihood of finding meaningful quantitative imaging biomarkers
-
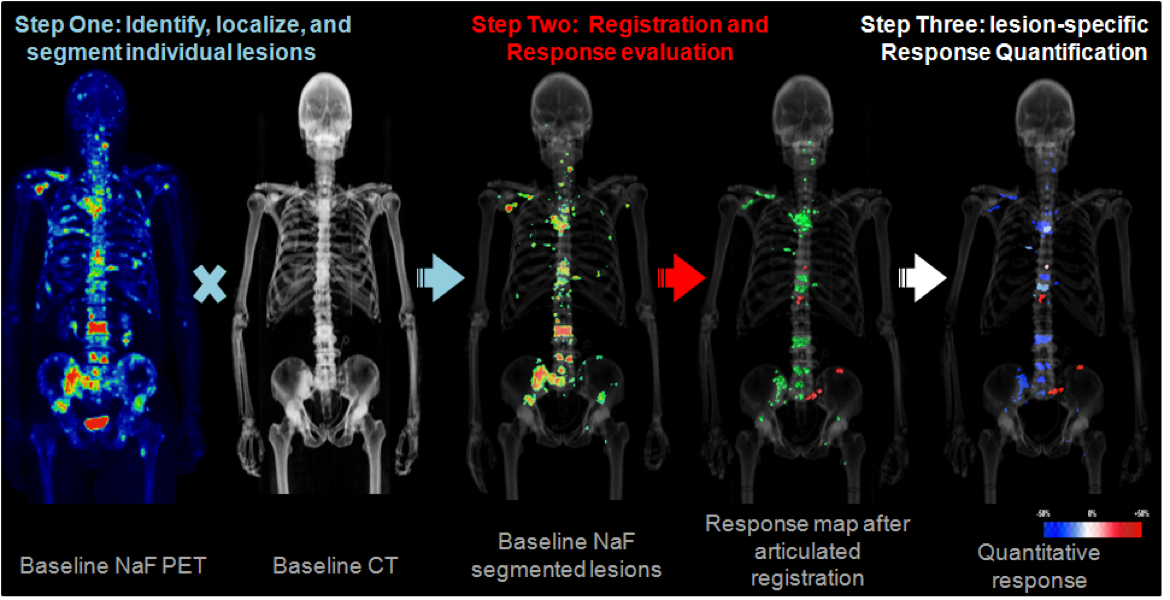
- Evaluation of metastatic patients often relies on whole-body biomarkers
- Semi-quantitative biomarkers are of limited use for clinical decisions
- Currently developing technique to track individual lesions during therapy to extract quantitative imaging data for treatment response evaluation
Current research focuses:
- Automatic identification and segmentation of metastatic vs. degenerative joint disease
- Identification of imaging features associated with biological resistance mechanisms
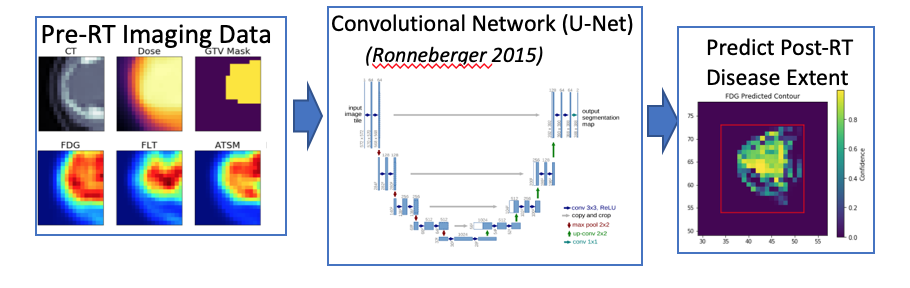
- Prediction of post-treatment disease extent from molecular imaging and radiotherapy planning data
- Representation learning-based segmentation of abdominal organs for off-target PET monitoring
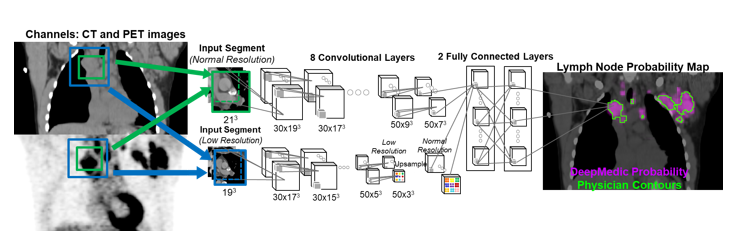
- Detection of lymph nodes for automated lymphoma response assessment
- PET image synthesis from CT
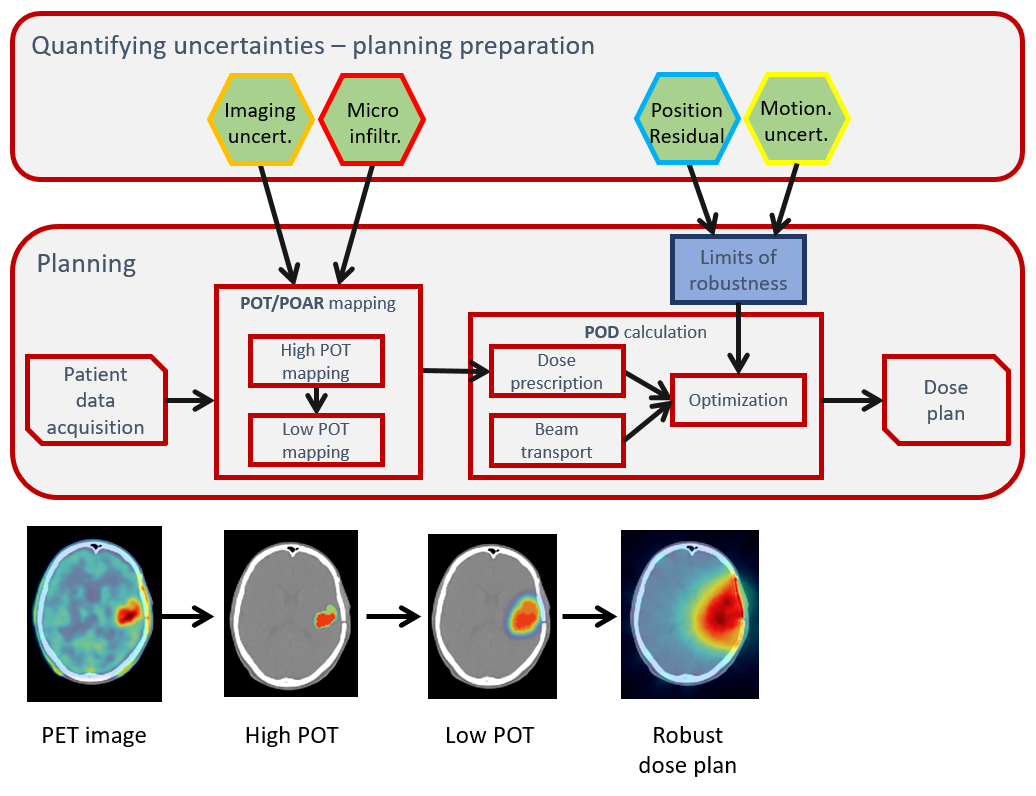
- The ICRU Reports 50, 62 and 83, which have established precise terminology to describe different areas of tumor presence, define current RT clinical paradigm through the use of GTV/CTV/PTV and expansions of volumes by margins.
- Although such approach is pragmatic and historically sensible, simply adding margins in such a linear way is a method that limits the implementation of non-uniform dose techniques, such as dose painting by the numbers